|
Effects of ECM protein micropatterns on the migration and differentiation of adult neural stem cells
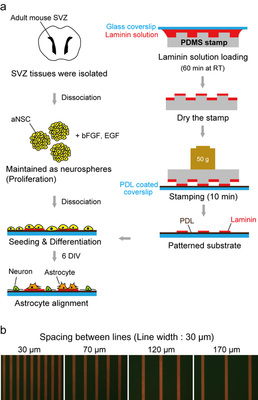
Sunghoon Joo, Joo Yeon Kim, Eunsoo Lee, Nari Hong, Woong Sun & Yoonkey Nam This is an outcome of our on-going collaboration with Prof. Woong Sun's lab in Korea University (고려대학교 해부학 교실, 선웅 교수). In this work, Sunghoon (KAIST, NEL) and Joo Yeon (Korea University College of Medicine) applied soft-lithographic technique to design simple and reproducible laminin (LN)-polylysine cell culture substrates and investigated how aNSCs respond to the various spatial distribution of laminin, one of ECM proteins enriched in the aNSC niche. They found that aNSC preferred to migrate and attach to LN stripes, and aNSC-derived neurons and astrocytes showed significant difference in motility towards LN stripes. By changing the spacing of LN stripes, they discovered a new way of separating neurons and astrocytes differentiated from adult neural stem cells. To the best of our knowledge, this is the first time to investigate the differential cellular responses of aNSCs on ECM protein (LN) and cell adhesive synthetic polymer (PDL) using surface micropatterns. NeuroCa: Integrated framework for systematic analysis of spatio-temporal neuronal activity patterns from large-scale optical recording data
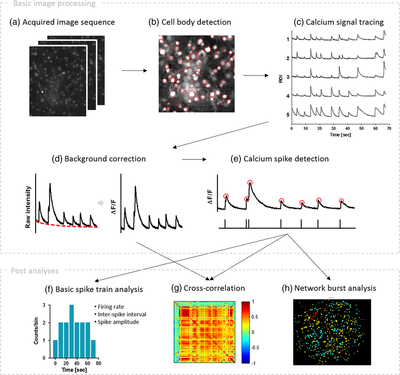
Minjee Jang and Yoonkey Nam In this work, Minjee developed a MATLAB-based toolbox, named NeuroCa, for the automated processing and quantitative analysis of large-scale calcium imaging data. This tool includes several computational algorithms to extract the calcium spike trains of individual neurons from the calcium imaging data in an automatic fashion. Two algorithms were developed to decompose the imaging data into the activity of individual cells and subsequently detect calcium spikes from each neuronal signal. Applying our method to dense networks in dissociated cultures, we were able to obtain the calcium spike trains of ∼1000 neurons in a few minutes. Further analyses using these data permitted the quantification of neuronal responses to chemical stimuli as well as functional mapping of spatiotemporal patterns in neuronal firing within the spontaneous, synchronous activity of a large network. These results demonstrate that our method not only automates time-consuming, labor-intensive tasks in the analysis of neural data obtained using optical recording techniques but also provides a systematic way to visualize and quantify the collective dynamics of a network in terms of its cellular elements. For more information please go to the NeuroCa webpage (link) |
Categories
All
Archives
December 2023
|
|
Korea Advanced Institute of Science and Technology (KAIST)
291 Daehak-ro, Yuseong-gu Daejeon 34141, Republic of Korea https://www.kaist.ac.kr Phone: +82-42-350-5362 |

 RSS Feed
RSS Feed
